sh <- suppressPackageStartupMessages
sh(library(tidyverse))
sh(library(caret))
sh(library(fastDummies))
wine <- readRDS(gzcon(url("https://github.com/cd-public/D505/raw/master/dat/wine.rds")))Abstract:
This is a technical blog post of both an HTML file and .qmd file hosted on GitHub pages.
Setup
Step Up Code:
Explanataion:
We will proceed as follows:
- Suppresslibraryload warnings, as we addressed them prior to publication.
- Use Tidy data sets via thetidyversepackage
- Perform classification and regression via thecaretpackage
- Engineer features via thefastDummiespackage
- Load in thewine.rdsdataframe, hosted publicly on GitHub.
Feature Engineering
We begin by engineering an number of features.
wino <- wine %>%
mutate(lprice=log(price), description = tolower(description)) %>%
select(lprice, description)- Create a total of 10 features (including points).
notes <- c("smoke", "spice", "pepper", "grass", "tannic", "crisp", "acidic", "bright", "smooth")
for (note in notes) {
wino <- wino %>%
mutate(!!sym(note) := str_detect(description, note))
}
head(wino)# A tibble: 6 × 11
lprice description smoke spice pepper grass tannic crisp acidic bright smooth
<dbl> <chr> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl>
1 2.71 this is rip… FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE
2 2.64 tart and sn… FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE
3 2.56 pineapple r… FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
4 4.17 much like t… FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE
5 2.71 blackberry … FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
6 2.77 here's a br… FALSE FALSE TRUE FALSE FALSE FALSE FALSE TRUE FALSE - Remove all rows with a missing value.
- Ensure only log(price) and engineering features are the only columns that remain in the
winodataframe.
wino <- wino %>% select(-description) %>% drop_na(.)Caret
We now use a train/test split to evaluate the features.
- Use the Caret library to partition the wino dataframe into an 80/20 split.
set.seed(505) # For reproducibility
trainIndex <- createDataPartition(wino$lprice, p = 0.8, list = FALSE)
trainData <- wino[trainIndex, ]
testData <- wino[-trainIndex, ]- Run a linear regression with bootstrap resampling.
options(warn=-1)
model <- train(lprice ~ ., data = trainData, method = "lm", trControl = trainControl(method = "boot", number = 5))
options(warn=0)- Report RMSE on the test partition of the data.
sqrt(mean((predict(model, newdata = testData) - testData$lprice)^2))[1] 0.6475555Variable selection
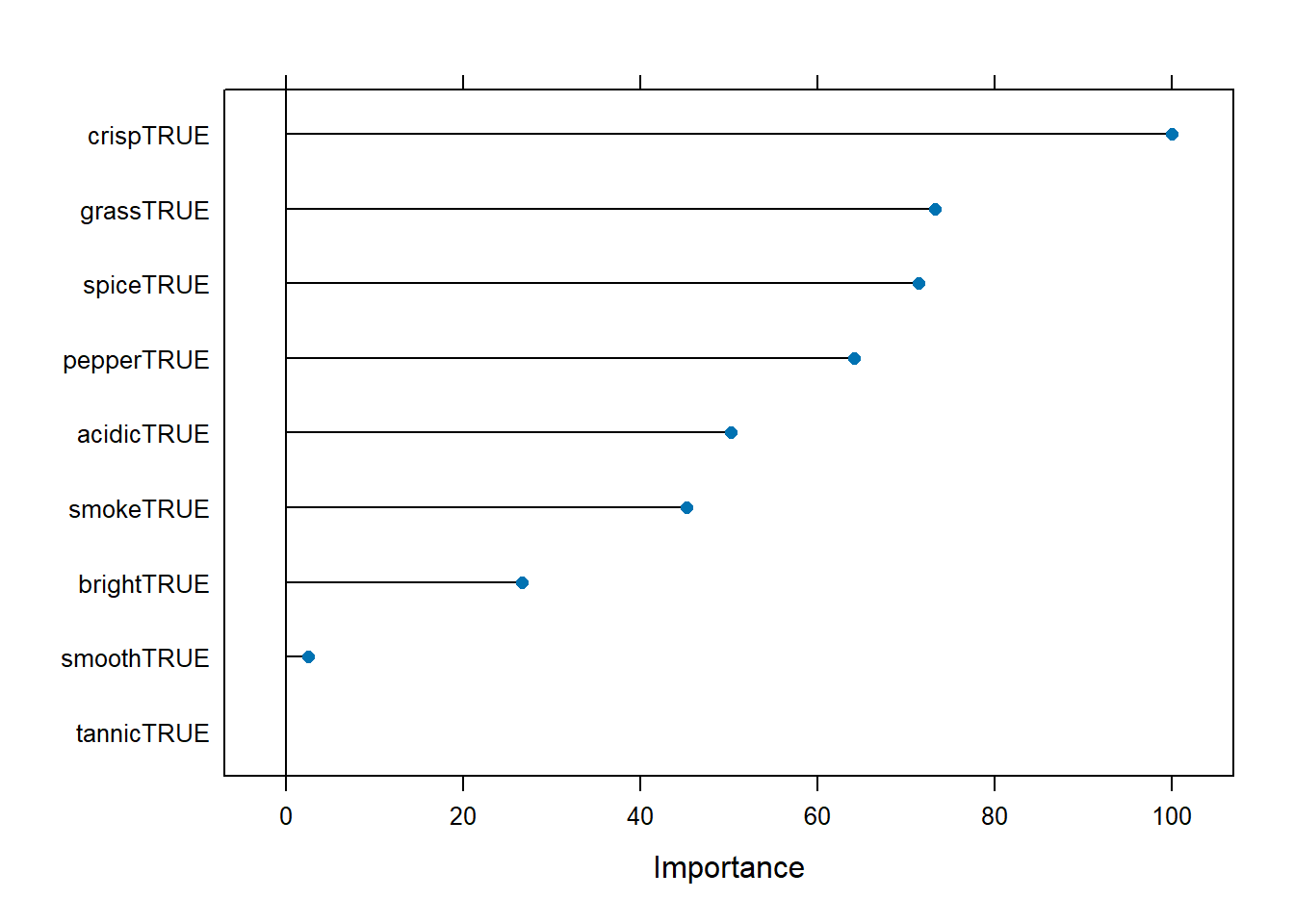
We now graph the importance of our 10 features.
plot(varImp(model))