Logistic Regression
Applied Machine Learning
Agenda
Agenda
- Course Announcements
- Mathematics of logistic regression
- Implementation with Caret
- ROC Curves
- Group work
Midterm 3/17
Brief recap of course so far
- Linear regression (e.g.
lprice ~ .), assumptions of model, interpretation. - \(K\)-NN (e.g.,
province ~ .), multi-class supervised classification. Hyperparameter \(k\). - Naive Bayes (e.g.,
province ~ .), multi-class supervised classification. - Logistic regression (e.g.,
province=="Oregon" ~ .), binary supervised classification. Elastic net. - Feature engineering (logarithms, center/scaling, Box Cox, tidytext, etc.).
- Feature selection (correlation, linear / logistic coefficients, frequent words, frequent words by class, etc.).
Practice
- I am working to develop a practice midterm.
- I will circulate it by 3 Mar.
- You will go over it on 10 Mar.
- It is based on the 5 homeworks.
- It is based on the prior slide.
- Little to no computatational linguistics
- I’m regarding
tidytextas extension, not core, content.
Modality Discussion
- I would release an assignment electronically at 6 PM
- We can do in person or otherwise.
- It will be “cheat proof”
- I will ask you nothing for which it matters how you determine the answer.
- If e.g. ChatGPT can be mind controlled into doing high quality feature engineering, you get points for mind controlling ChatGPT.
First Model Due 3/10
Publish
Each group should create: 1. An annotated .*md file, and 2. The .rds/.pickle/.parquet file that it generates, that 3. Contains only the features you want in the model.
Under version control, on GitHub.
Constraints
I will run:
- The specified \(K\)NN or Naive Bayes model,
- With:
province ~ .(or the whole data frame inscikit) - With repeated 5-fold cross validation
- With the same index for partitioning training and test sets for every group.
- On whatever is turned in before class.
- Bragging rights for highest Kappa
Context
- The “final exam” is that during the last class you will present your model results as though you are speaking to the managers of a large winery.
- It should be presented from a Quarto presentation on GitHub or perhaps e.g. RPubs.
- You must present via the in-room “teaching machine” computer, not your own physical device, to ensure that you are comfortable distributing your findings.
Group Meetings
- You should have a group assignment
- Meet in your groups!
- Talk about your homework with your group.
Logistic Regression
Algorithm
- Assume a linear relationship between the log odds and a set of predictor variables.
\[ log(\frac{p}{1-p})=\beta_{0}+\beta_{1}x_{1}+\beta_{2}x_{2} \]
With a bit of algebra you can get the probabilities as…
\[ p=\frac{1}{1+e^{-(\beta_{0}+\beta_{1}x_{1}+\beta_{2}x_{2})}} \]
\[ log(\frac{p}{1-p})=\beta_{0}+\beta_{1}x_{1}+\beta_{2}x_{2} \]
‘p’ represents the probability of the event occurring.
- Represented as natural log
ln… - of the odds ratio (\(\frac{p}{1-p}\))…
- as a linear combination of
- predictor variables (\(x_1\), \(x_2\)) and
- their corresponding coefficients (\(\beta_1\), \(\beta_2\))
- plus an intercept (\(\beta_0\)).
Understanding check?
- Why do we call this regression instead of classification?
- Quick Algebra review
- Look, it’s better to be comfortable with algebra than not.
Log Odds
- 1. Start with the log-odds equation:
\[ log(\frac{p}{1-p}) = \beta_0 + \beta_1x_1 + \beta_2x_2 \]
Exponentiate
2. Exponentiate both sides:
To remove the logarithm, we take the exponential (e) of both sides of the equation:
\[ e^{log(\frac{p}{1-p})} = e^{\beta_0 + \beta_1x_1 + \beta_2x_2} \]
Simplify
3. Simplify the left side:
The exponential function and the natural logarithm are inverse functions of each other.
Therefore, \(e^{log(x)} = x\). This simplifies the left side:
\[ \frac{p}{1-p} = e^{\beta_0 + \beta_1x_1 + \beta_2x_2} \]
Rewrite
4. Rewrite the right side using exponent rules:
We can rewrite the right side using the rule \(e^{a+b} = e^a * e^b\):
\[ \frac{p}{1-p} = e^{\beta_0} * e^{\beta_1x_1} * e^{\beta_2x_2} \]
Find \(p\) 1
- Find \(p\)
- Multiply both sides by (1-p):
\[ p = (1-p) * e^{\beta_0} * e^{\beta_1x_1} * e^{\beta_2x_2} \]
Find \(p\) 2
- Find \(p\)
- Distribute the right-hand side (RHS):
\[ p = e^{\beta_0} * e^{\beta_1x_1} * e^{\beta_2x_2} - p * e^{\beta_0} * e^{\beta_1x_1} * e^{\beta_2x_2} \]
Find \(p\) 2
- Find \(p\)
- Move the \(p\) term to the left side:
\[ p + p * e^{\beta_0} * e^{\beta_1x_1} * e^{\beta_2x_2} = e^{\beta_0} * e^{\beta_1x_1} * e^{\beta_2x_2} \]
Find \(p\) 3
- Find \(p\)
- Factor out \(p\) on the left side:
\[ p * (1 + e^{\beta_0} * e^{\beta_1x_1} * e^{\beta_2x_2}) = e^{\beta_0} * e^{\beta_1x_1} * e^{\beta_2x_2} \]
Find \(p\) 4
- Find \(p\)
- Divide both sides by the term in parentheses: \[ p = \frac{e^{\beta_0} * e^{\beta_1x_1} * e^{\beta_2x_2}}{1 + e^{\beta_0} * e^{\beta_1x_1} * e^{\beta_2x_2}} \]
Simplify
6. Simplify the expression:
We can further simplify by multiplying the numerator and denominator by \(e^{-(\beta_0 + \beta_1x_1 + \beta_2x_2)}\):
\[ p = \frac{1}{e^{-(\beta_0 + \beta_1x_1 + \beta_2x_2)} + 1} \]
Rewrite
- Which is commonly written as:
\[ p = \frac{1}{1 + e^{-(\beta_0 + \beta_1x_1 + \beta_2x_2)}} \]
Implementation with Caret
Libraries Setup
Dataframe
Extract certain words
- We’ll proceed in a few steps.
- First, a function to extract all words.
- We’ll omit stopwords
- Remember those? Like “of” and “a”
- We’ll omit a few words-of-choice
- Like “pinot”
- We’ll omit stopwords
- First, a function to extract all words.
Extract all words
Extract all words
words <- desc_to_words(wine, c("wine","pinot","vineyard"))
# The second argument is our custom stopwards, as a vector
head(words) id province price points year word
1 1 Oregon 65 87 2012 regular
2 1 Oregon 65 87 2012 bottling
3 1 Oregon 65 87 2012 2012
4 1 Oregon 65 87 2012 rough
5 1 Oregon 65 87 2012 tannic
6 1 Oregon 65 87 2012 rusticExtract certain words
- We’ll proceed in a few steps.
- Optionally, look at stems
- This is really cool.
- Do I care about the difference between “acidic” and “acidity”?
- The linguistics consensus leans toward no.
- Optionally, look at stems
STEM
- Short for data Science, daTa science, data sciencE, and Machine learning.
- We want to use a
tidytextbuilt-in here, of course.- We aren’t domain experts in linguistics…
- But we are domain experts in using R libraries.
Example
Wait
- ‘wait’ (infinitive, imperative, present subjunctive, and present indicative except in the 3rd-person singular)
- ‘waits’ (3rd person singular simple present indicative)
- ‘waited’ (simple past)
- ‘waited’ (past participle)
- ‘waiting’ (present participle)
STEM
Bottl
- Another example.
Like a G6
Word Count
- With either or words or our stems, we can see how many of each word we have easily enough.
- And eliminate words will less than a certain count!
Check it
Back to Wine
- We currently have multiple entries per ID in a tidy dataframe
- We would like to get back to have features for each ID, as each ID is some wine.
- One way to do so, is to make columns from the word data.
- We use
pivot_wider
Pivot
Check it
# A tibble: 6 × 23
drink oak aromas bodied raspberry spice finish fruit flavors tannins
<lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl>
1 TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
2 FALSE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE
3 FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE FALSE FALSE
4 FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE
5 TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE
6 FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE
# ℹ 13 more variables: strawberry <lgl>, cranberry <lgl>, dark <lgl>,
# palate <lgl>, acidity <lgl>, black <lgl>, cherry <lgl>, nose <lgl>,
# ripe <lgl>, light <lgl>, red <lgl>, plum <lgl>, province <chr># A tibble: 6 × 30
bottl ag berri drink oak aroma bodi offer raspberri spice textur finish
<lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl>
1 TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
2 FALSE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
3 FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE
4 FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE
5 FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
6 FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
# ℹ 18 more variables: flavor <lgl>, fruit <lgl>, ripe <lgl>, tannin <lgl>,
# strawberri <lgl>, cranberri <lgl>, dark <lgl>, palat <lgl>, acid <lgl>,
# black <lgl>, cherri <lgl>, nose <lgl>, light <lgl>, red <lgl>, rich <lgl>,
# plum <lgl>, structur <lgl>, province <chr>Full Function
- Create a function to extract words with totals > j
Look at the data
# A tibble: 10 × 6
drink oak aromas bodied raspberry province
<lgl> <lgl> <lgl> <lgl> <lgl> <chr>
1 TRUE TRUE FALSE FALSE FALSE Oregon
2 FALSE TRUE TRUE TRUE TRUE California
3 FALSE FALSE FALSE FALSE FALSE Oregon
4 FALSE FALSE FALSE FALSE FALSE Oregon
5 TRUE FALSE FALSE FALSE FALSE Oregon
6 FALSE FALSE FALSE FALSE TRUE California
7 FALSE FALSE FALSE FALSE TRUE California
8 FALSE TRUE FALSE FALSE TRUE California
9 FALSE FALSE FALSE FALSE FALSE Oregon
10 FALSE FALSE TRUE FALSE FALSE Oregon Logistic Regression
- We check a true/false outcome.
wino <- wino %>%
mutate(oregon = factor(province=="Oregon")) %>%
select(-province)
wino %>%
head(10) %>%
select(1:5, oregon)# A tibble: 10 × 6
drink oak aromas bodied raspberry oregon
<lgl> <lgl> <lgl> <lgl> <lgl> <fct>
1 TRUE TRUE FALSE FALSE FALSE TRUE
2 FALSE TRUE TRUE TRUE TRUE FALSE
3 FALSE FALSE FALSE FALSE FALSE TRUE
4 FALSE FALSE FALSE FALSE FALSE TRUE
5 TRUE FALSE FALSE FALSE FALSE TRUE
6 FALSE FALSE FALSE FALSE TRUE FALSE
7 FALSE FALSE FALSE FALSE TRUE FALSE
8 FALSE TRUE FALSE FALSE TRUE FALSE
9 FALSE FALSE FALSE FALSE FALSE TRUE
10 FALSE FALSE TRUE FALSE FALSE TRUE Split the data
A basic model
Check Kappa
Penalized Multinomial Regression
6644 samples
22 predictor
6 classes: 'Burgundy', 'California', 'Casablanca_Valley', 'Marlborough', 'New_York', 'Oregon'
No pre-processing
Resampling: Cross-Validated (5 fold)
Summary of sample sizes: 5315, 5315, 5315, 5316, 5315
Resampling results across tuning parameters:
decay Accuracy Kappa
0e+00 0.7245610 0.5683360
1e-04 0.7245610 0.5683360
1e-01 0.7251632 0.5691676
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was decay = 0.1.Probability
- See top coefficients
get_odds <- function(fit) {
as.data.frame(t(exp(coef(fit$finalModel)))) %>%
rownames_to_column(var = "name") %>%
pivot_longer(-name, names_to = "class", values_to = "odds") %>%
arrange(desc(odds)) %>%
head()
}
get_odds(fit)# A tibble: 6 × 3
name class odds
<chr> <chr> <dbl>
1 bodiedTRUE California 5.19
2 (Intercept) Oregon 3.92
3 noseTRUE California 2.80
4 oakTRUE California 2.65
5 aromasTRUE California 2.63
6 aromasTRUE Casablanca_Valley 2.30Confusion Matrix
get_matrix <- function(fit, df) {
pred <- factor(predict(fit, newdata = df))
confusionMatrix(pred,factor(df$oregon))
}
get_matrix(fit,test)Confusion Matrix and Statistics
Reference
Prediction FALSE TRUE
FALSE 996 165
TRUE 125 373
Accuracy : 0.8252
95% CI : (0.806, 0.8432)
No Information Rate : 0.6757
P-Value [Acc > NIR] : < 2e-16
Kappa : 0.5933
Mcnemar's Test P-Value : 0.02201
Sensitivity : 0.8885
Specificity : 0.6933
Pos Pred Value : 0.8579
Neg Pred Value : 0.7490
Prevalence : 0.6757
Detection Rate : 0.6004
Detection Prevalence : 0.6998
Balanced Accuracy : 0.7909
'Positive' Class : FALSE
Not bad. But what if we decrease threshold for a word to be included?
Using more words
Check Kappa
Penalized Multinomial Regression
6696 samples
54 predictor
6 classes: 'Burgundy', 'California', 'Casablanca_Valley', 'Marlborough', 'New_York', 'Oregon'
No pre-processing
Resampling: Cross-Validated (5 fold)
Summary of sample sizes: 5359, 5356, 5356, 5357, 5356
Resampling results across tuning parameters:
decay Accuracy Kappa
0e+00 0.744476 0.6200617
1e-04 0.744476 0.6200617
1e-01 0.743879 0.6192105
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was decay = 1e-04.Check odds
Confusion Matrix
Confusion Matrix and Statistics
Reference
Prediction Burgundy California Casablanca_Valley Marlborough New_York
Burgundy 188 16 2 2 4
California 29 617 8 16 9
Casablanca_Valley 0 9 10 0 1
Marlborough 0 1 0 1 0
New_York 0 1 0 0 4
Oregon 21 146 6 26 8
Reference
Prediction Oregon
Burgundy 40
California 70
Casablanca_Valley 1
Marlborough 0
New_York 1
Oregon 434
Overall Statistics
Accuracy : 0.7504
95% CI : (0.729, 0.771)
No Information Rate : 0.4728
P-Value [Acc > NIR] : < 2.2e-16
Kappa : 0.6107
Mcnemar's Test P-Value : NA
Statistics by Class:
Class: Burgundy Class: California Class: Casablanca_Valley
Sensitivity 0.7899 0.7810 0.384615
Specificity 0.9553 0.8502 0.993313
Pos Pred Value 0.7460 0.8238 0.476190
Neg Pred Value 0.9648 0.8124 0.990303
Prevalence 0.1424 0.4728 0.015560
Detection Rate 0.1125 0.3692 0.005984
Detection Prevalence 0.1508 0.4482 0.012567
Balanced Accuracy 0.8726 0.8156 0.688964
Class: Marlborough Class: New_York Class: Oregon
Sensitivity 0.0222222 0.153846 0.7949
Specificity 0.9993850 0.998784 0.8160
Pos Pred Value 0.5000000 0.666667 0.6771
Neg Pred Value 0.9736369 0.986787 0.8913
Prevalence 0.0269300 0.015560 0.3268
Detection Rate 0.0005984 0.002394 0.2597
Detection Prevalence 0.0011969 0.003591 0.3836
Balanced Accuracy 0.5108036 0.576315 0.8054Using stems
Check Kappa
Penalized Multinomial Regression
6689 samples
29 predictor
6 classes: 'Burgundy', 'California', 'Casablanca_Valley', 'Marlborough', 'New_York', 'Oregon'
No pre-processing
Resampling: Cross-Validated (5 fold)
Summary of sample sizes: 5351, 5351, 5353, 5350, 5351
Resampling results across tuning parameters:
decay Accuracy Kappa
0e+00 0.744509 0.5974314
1e-04 0.744509 0.5974314
1e-01 0.744509 0.5974314
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was decay = 0.1.Check odds
Confusion Matrix
Confusion Matrix and Statistics
Reference
Prediction Burgundy California Casablanca_Valley Marlborough New_York
Burgundy 181 13 1 3 7
California 40 597 8 16 10
Casablanca_Valley 0 2 4 0 1
Marlborough 0 0 0 0 0
New_York 0 0 0 1 0
Oregon 17 178 13 25 8
Reference
Prediction Oregon
Burgundy 28
California 83
Casablanca_Valley 0
Marlborough 0
New_York 0
Oregon 434
Overall Statistics
Accuracy : 0.7281
95% CI : (0.7061, 0.7494)
No Information Rate : 0.4731
P-Value [Acc > NIR] : < 2.2e-16
Kappa : 0.5716
Mcnemar's Test P-Value : NA
Statistics by Class:
Class: Burgundy Class: California Class: Casablanca_Valley
Sensitivity 0.7605 0.7557 0.153846
Specificity 0.9637 0.8216 0.998175
Pos Pred Value 0.7768 0.7918 0.571429
Neg Pred Value 0.9603 0.7893 0.986771
Prevalence 0.1425 0.4731 0.015569
Detection Rate 0.1084 0.3575 0.002395
Detection Prevalence 0.1395 0.4515 0.004192
Balanced Accuracy 0.8621 0.7886 0.576011
Class: Marlborough Class: New_York Class: Oregon
Sensitivity 0.00000 0.0000000 0.7963
Specificity 1.00000 0.9993917 0.7858
Pos Pred Value NaN 0.0000000 0.6430
Neg Pred Value 0.97305 0.9844218 0.8884
Prevalence 0.02695 0.0155689 0.3263
Detection Rate 0.00000 0.0000000 0.2599
Detection Prevalence 0.00000 0.0005988 0.4042
Balanced Accuracy 0.50000 0.4996959 0.7911Even better?
Receiver Operating Characteristic (ROC) Curve
ROC Curve
Image

Sensitivity
- Sensitivity: True Positive Rate
- Measures how well the model detects true positives.
- TP: True Positive
- FN: False Negative
\[ \text{Sensitivity} = \frac{\text{TP}}{\text{TP} + \text{FN}} \]
Specificifity
- Specificity: True Negative Rate
- Measures how well the model avoids false positives.
- FP: False Positive
- TN: True Negative
\[ \text{Specificity} = \frac{\text{TN}}{\text{TN} + \text{FP}} \]
Storm Prediction
- True Positive (TP): The model predicts a storm, and a storm actually occurs.
- False Positive (FP): The model predicts a storm, but no storm occurs (false alarm).
- True Negative (TN): The model predicts no storm, and no storm occurs.
- False Negative (FN): The model predicts no storm, but a storm actually occurs (missed event).
Goals
- A perfect model would have both
- high sensitivity
- (detecting all real storms) and
- high specificity
- (avoiding false alarms).
- high sensitivity
- The ROC curve helps analyze this trade-off.
Interpreting the ROC Curve
- A point near (0,1) on the ROC curve represents high sensitivity and high specificity (ideal performance).
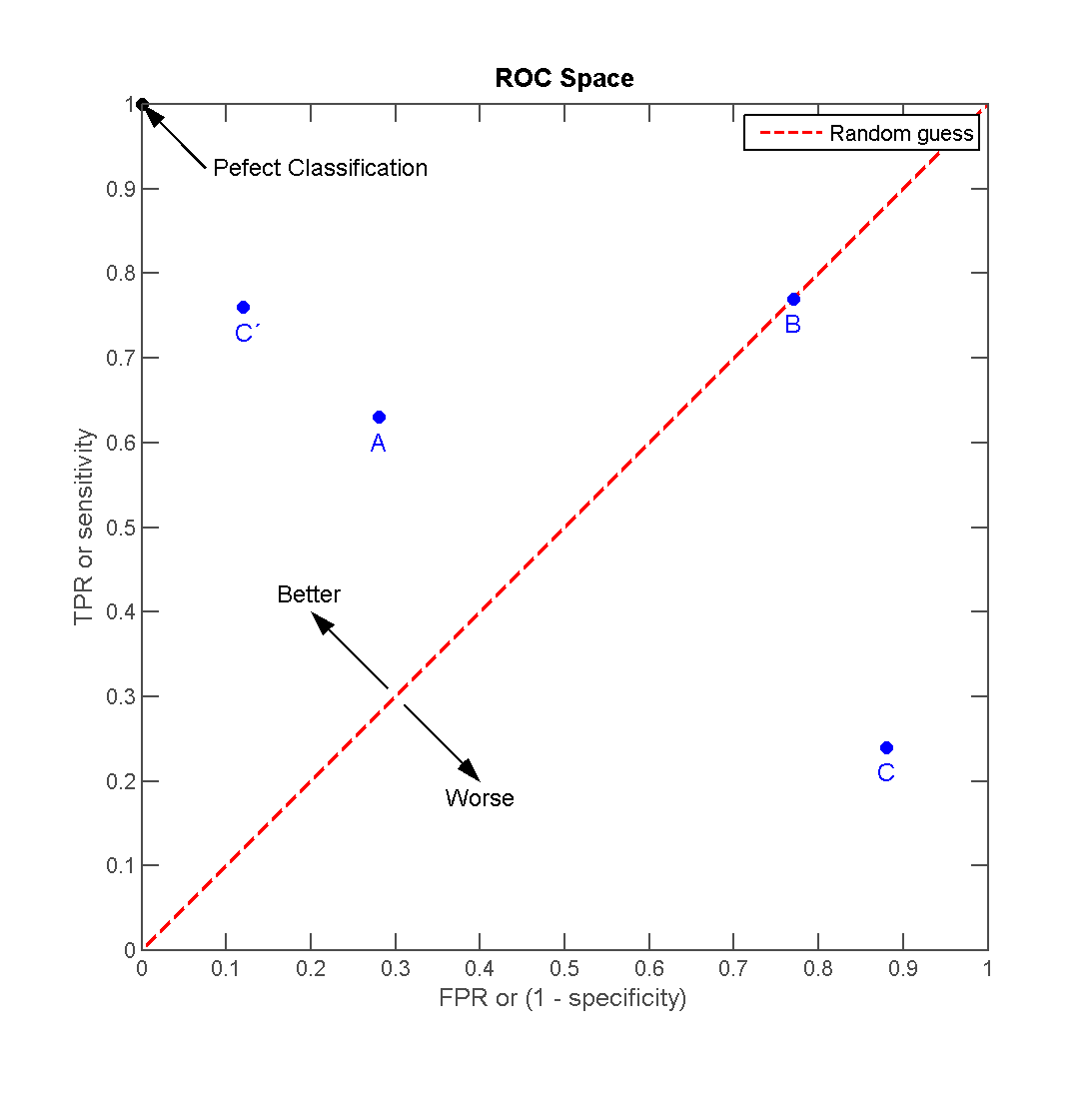
Interpreting the ROC Curve
- A curve closer to the diagonal line (random guessing) indicates poor predictive ability.

Interpreting the ROC Curve
- The Area Under the Curve (AUC) quantifies the overall model performance, with AUC = 1 being perfect and AUC = 0.5 being no better than chance.
Model Tuning
- In e.g. a storm model….
- adjusting the model’s decision threshold
- (e.g., how strong a weather signal needs to be before predicting a storm),
- we can move along the ROC curve to balance
- sensitivity and
- specificity.
ROC Curve evaluation
- Let’s look at our most recent fit:
See the curve
Area under the curve: 0.9175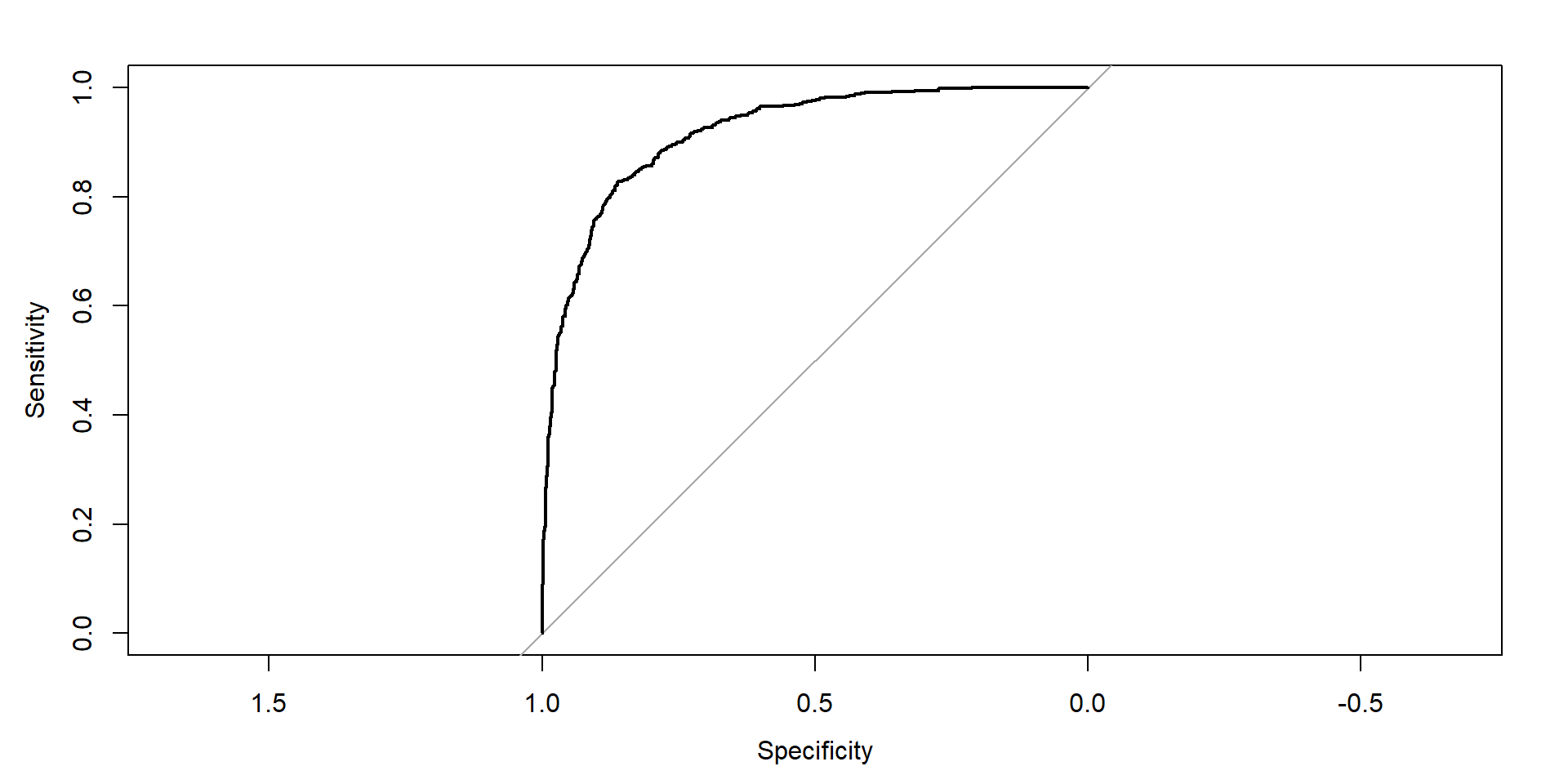
Exercise
- Gather into your prediction teams.
- Choose a Pinot province other than Oregon or California
- Use logistic regression to find the words/terms that increase the odds of choosing that province the most
Vocabulary
- Logistic Regression
- Odds
- Sensitivity
- Specificity
- ROC
- AUC
