sh <- suppressPackageStartupMessages
sh(library(tidyverse))
sh(library(tidytext))
sh(library(caret))
sh(library(topicmodels)) # new?
data(stop_words)
sh(library(thematic))
theme_set(theme_dark())
thematic_rmd(bg = "#111", fg = "#eee", accent = "#eee")
wine <- readRDS(gzcon(url("https://cd-public.github.io/D505/dat/variety.rds"))) %>% rowid_to_column("id")
bank <- readRDS(gzcon(url("https://cd-public.github.io/D505/dat/BankChurners.rds")))Dimensionality Reduction
Applied Machine Learning
Agenda
- Review of Mid-term
- Next modeling project
- Principle component analysis
- Topic modeling
- Group breakout sessions (if time)
Timing Update
- I misnumbered my weeks, so Model 2 is due immediately before the final.
- In theory, this means you have more time on it.
- Anyways:
- Model 2 by 14 Apr
- Final on 21 Apr
Next modeling project
- Due 2 more Mondays (14 Apr)
- Use bank data to predict churn
- Using only 5 features
- Models scored based on AUC
- We’ll leave time at the end to get started
Principle component analysis
How?
Linear Transform
- We use this notion of linear combinations a lot:
- Features with coeffiencients
- Street address
- Course numbers
- DATA 505 is a combination of
- a non-numerical (categorical) prefix
- a “hundred level” (5, for MS) and
- a course number (05)
Non-independence
- There are many more 100 and 200 level courses than 500
- So we shouldn’t assume that “05” means the same thing after “5” as it does after “1”
- Many prefixes \(\times\) hundred levels lack an “01” course at all!
- So we shouldn’t assume that “05” means the same thing after “5” as it does after “1”
Visualize
PCA of a multivariate Gaussian distribution centered at (1,3) with a standard deviation of 3 in roughly the (0.866, 0.5) direction and of 1 in the orthogonal direction. The vectors shown are the eigenvectors of the covariance matrix scaled by the square root of the corresponding eigenvalue, and shifted so their tails are at the mean.
Why?
What are the primary reasons to use PCA?
- Dimensionality reduction
- Visualization
- Noise reduction
Curse of dimensionality
- As the dimensionality of the feature space increases,
- the number of configurations can grow exponentially, and thus
- the number of configurations covered by an observation decreases.
- Another formulation:
- distances between observations
- tend to shrink as
- dimensionality tends to infinity.
- distances between observations


Idea
- Find a linear combination of variables to create principle components
- Maintain as much variance as possible.
- Principle components are orthogonal (uncorrelated)
Rotation of orthogonal axes
PCA of a multivariate Gaussian distribution centered at (1,3) with a standard deviation of 3 in roughly the (0.866, 0.5) direction and of 1 in the orthogonal direction. The vectors shown are the eigenvectors of the covariance matrix scaled by the square root of the corresponding eigenvalue, and shifted so their tails are at the mean.
Singular value decomposition
Illustration of the singular value decomposition \(U\Sigma V⁎\) of matrix \(M\).
- Top: The action of \(M\), indicated by its effect on the unit disc \(D\) and the two canonical unit vectors \(e_1\) and \(e_2\).
- Left: The action of \(V⁎\), a rotation, on \(D\), \(e_1\) and \(e_2\).
- Bottom: The action of \(\Sigma\), a scaling by the singular values \(\sigma_1\) horizontally and \(\sigma_1\) vertically.
- Right: The action of \(U\), another rotation.
Libraries Setup
- Note we are putting
idinto the wine dataframe!- Why do we do this?
Wine Words
- Let’s take some wine words.
winewords <- function(df, cutoff) {
df %>%
unnest_tokens(word, description) %>% anti_join(stop_words) %>%
filter(!(word %in% c("drink","vineyard","variety","price","points","wine","pinot","chardonnay","gris","noir","riesling","syrah"))) %>%
count(id, word) %>% group_by(word) %>% mutate(total = sum(n)) %>% filter(total > cutoff) %>%
ungroup() %>% group_by(id) %>% mutate(exists = if_else(n>0,1,0)) %>% ungroup() %>%
pivot_wider(id_cols=id, names_from=word, values_from=exists, values_fill=c(exists=0)) %>%
right_join(wine, by="id") %>% drop_na(.) %>% mutate(log_price = log(price)) %>%
select(-id, -price, -description)
}Wine Words
- Let’s take some wine words.
PCA
prcomp: Principal Components Analysis
Performs a principal components analysis on the given data matrix and returns the results as an object of class prcomp.
PCA the wine
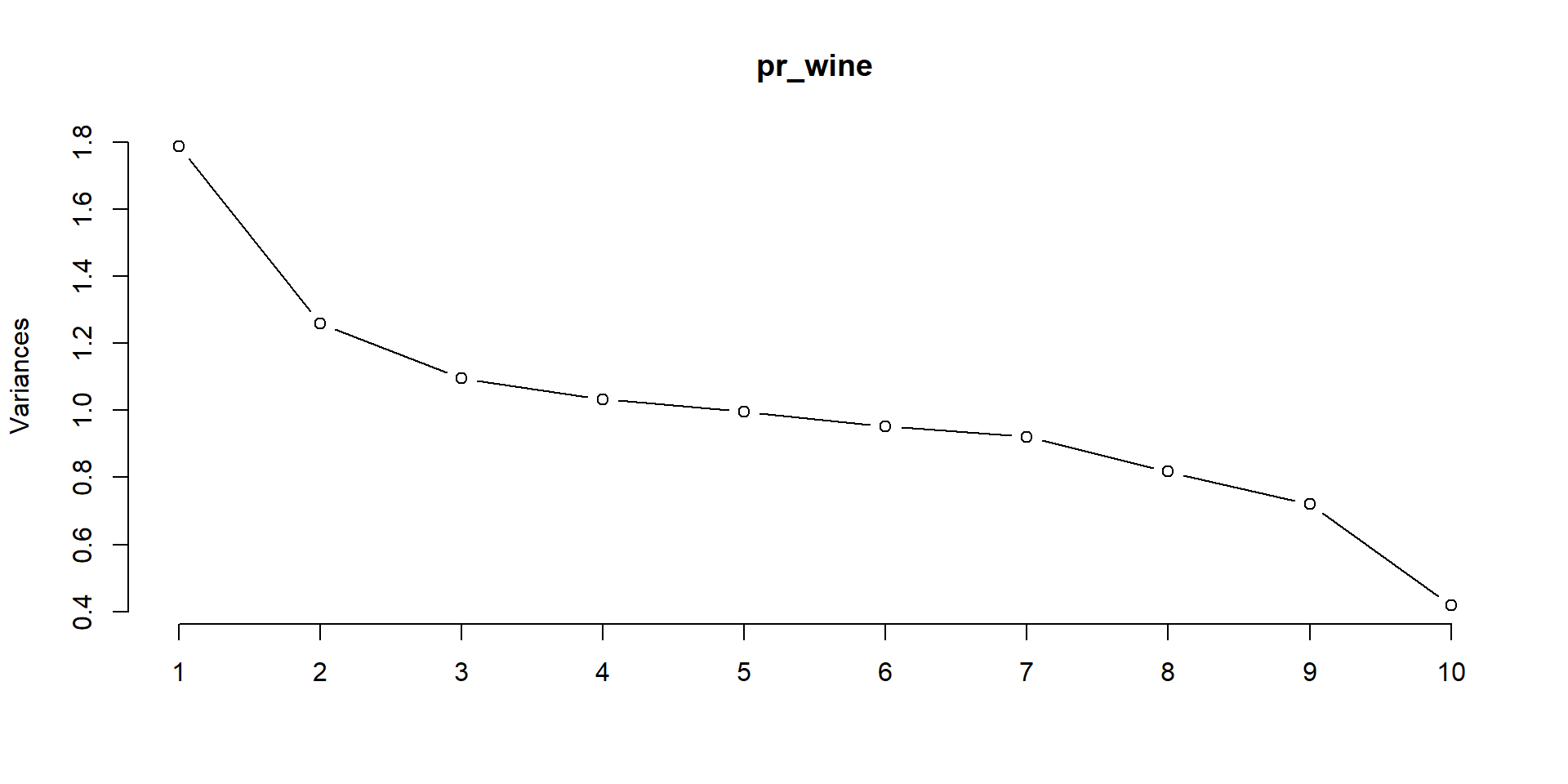
Importance of components:
PC1 PC2 PC3 PC4 PC5 PC6 PC7
Standard deviation 1.3365 1.1225 1.0462 1.0164 0.99785 0.97618 0.95977
Proportion of Variance 0.1786 0.1260 0.1095 0.1033 0.09957 0.09529 0.09212
Cumulative Proportion 0.1786 0.3046 0.4141 0.5174 0.61697 0.71226 0.80438
PC8 PC9 PC10
Standard deviation 0.90424 0.84859 0.64691
Proportion of Variance 0.08176 0.07201 0.04185
Cumulative Proportion 0.88614 0.95815 1.00000Show variance plot
Visualize biplots
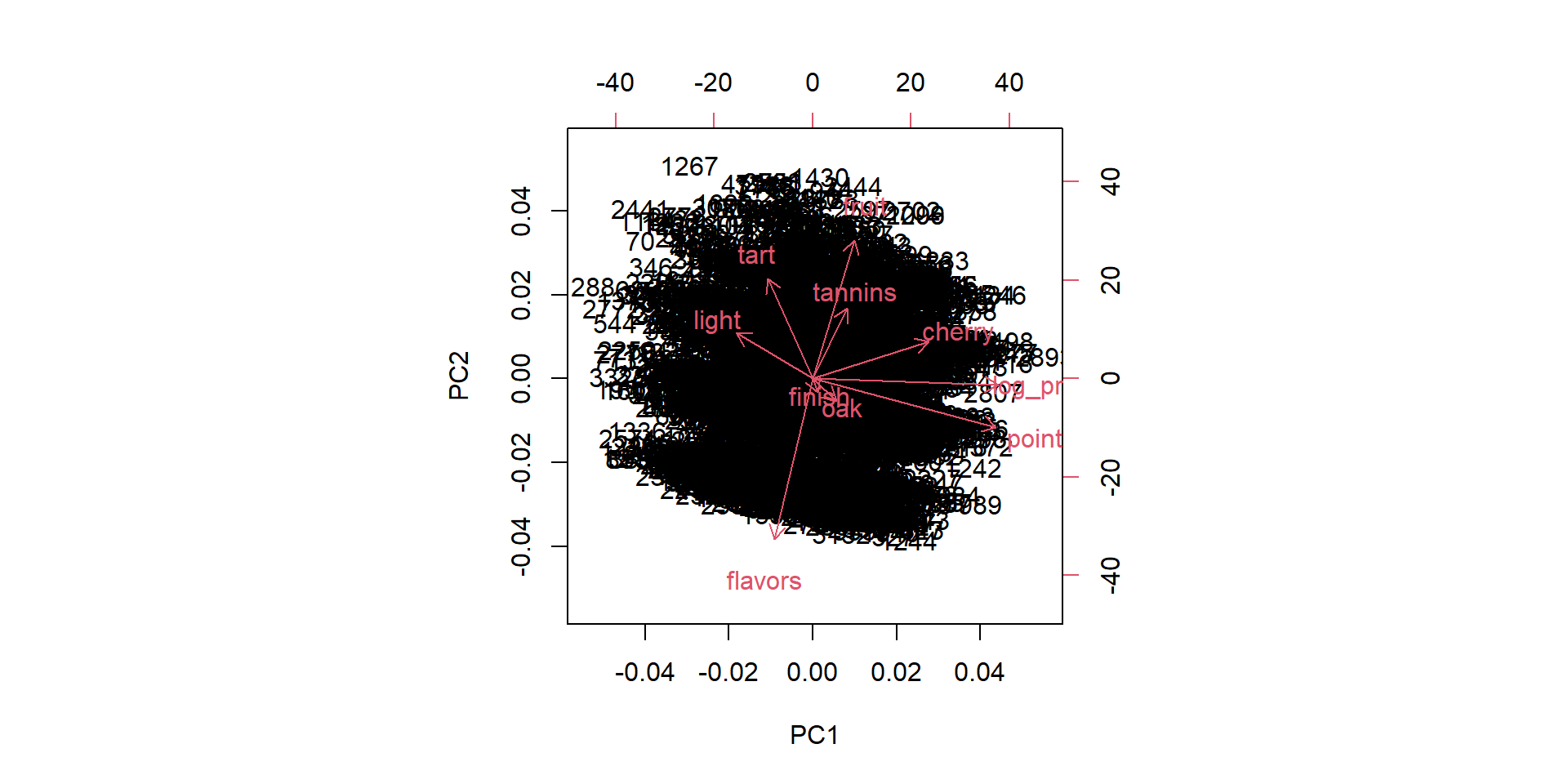
Visualize biplots
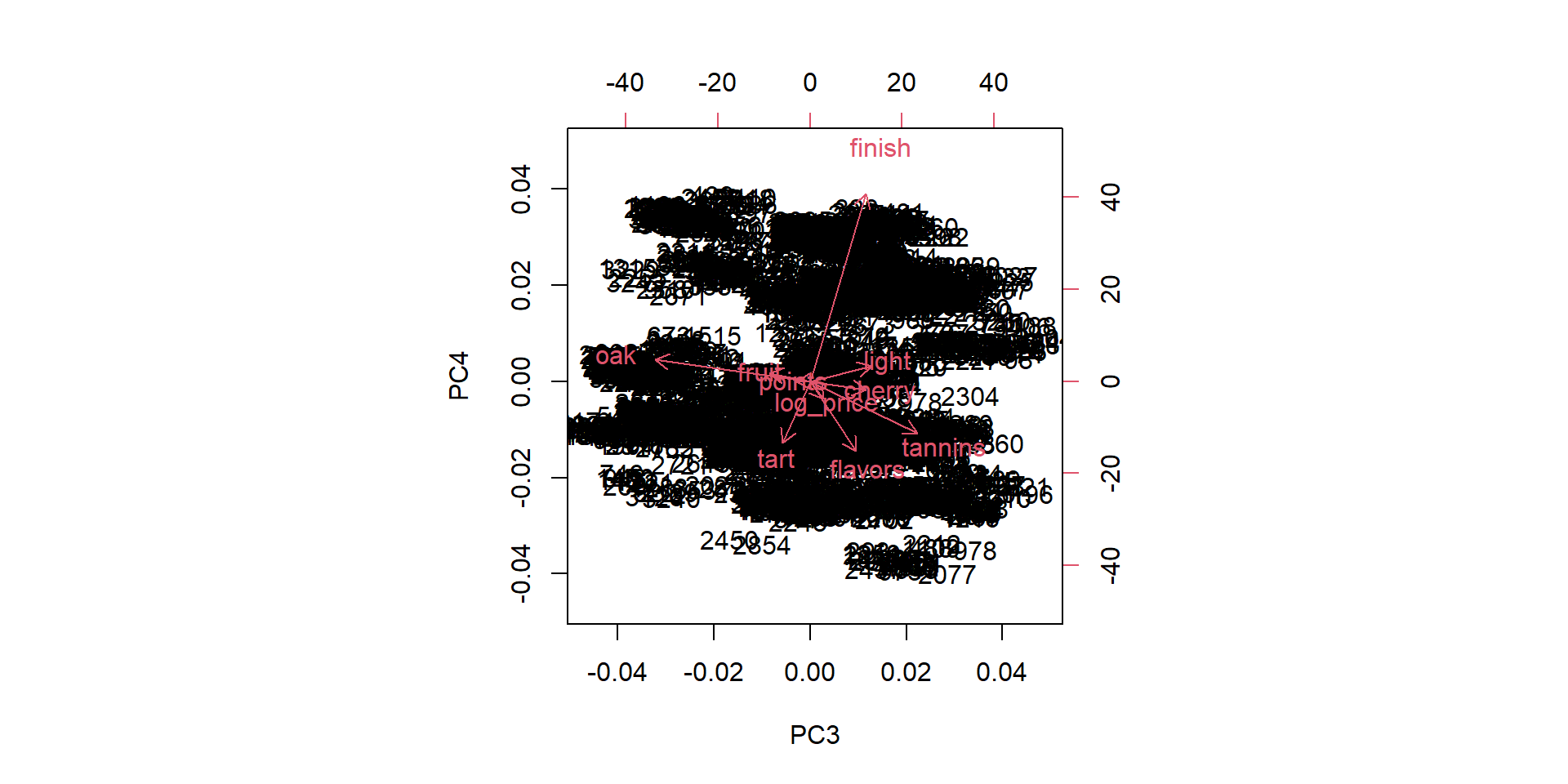
Factor loadings
PC1 PC2 PC3 PC4 PC5
flavors -0.12478797 -0.62231667 0.20452230 -0.318630695 0.13664856
tart -0.14685952 0.38830520 -0.12183008 -0.280795083 -0.21656122
oak 0.07633205 -0.08679216 -0.68973197 0.099583899 -0.47869174
finish 0.01746794 -0.05286174 0.25007481 0.859753722 -0.07885236
fruit 0.13660189 0.53749981 -0.17969818 0.037421478 0.49610389
tannins 0.11111542 0.27285155 0.47849402 -0.237346398 -0.40926470
cherry 0.37873576 0.14326980 0.24907181 -0.037509702 0.19615664
light -0.24837285 0.17747080 0.27319253 0.069367578 -0.43069001
points 0.59658742 -0.18887826 -0.05944313 -0.003297401 -0.05847348
log_price 0.60388362 -0.02372061 0.06006730 -0.081066609 -0.24095187
PC6 PC7 PC8 PC9 PC10
flavors -0.04766056 0.05424377 0.11086176 -0.64444967 0.04798526
tart -0.73492763 0.21914758 -0.15739332 -0.23651461 0.13493716
oak 0.29179417 -0.11495294 -0.19800128 -0.36140534 0.06059095
finish -0.29384173 -0.08875132 -0.03526432 -0.30446445 0.02840786
fruit 0.18035875 -0.04512780 0.40587621 -0.46049057 0.03563910
tannins 0.13904469 -0.62839693 0.01867847 -0.15124842 0.15158122
cherry 0.24157483 0.30605898 -0.73331841 -0.18300560 0.10408832
light 0.36764429 0.63115133 0.29992812 -0.07632032 0.11215674
points -0.14532219 0.11863618 0.29334800 0.15866137 0.67455570
log_price -0.14007525 0.15787054 0.20889886 -0.07756363 -0.68725919Plotly
library(plotly)
biplot_data <- data.frame(pr_wine$x)
biplot_data$variety <- wino$variety
p <- plot_ly(biplot_data, x = ~PC1, y = ~PC2, z = ~PC3, color = ~PC4, type = 'scatter3d', mode = 'markers') %>%
layout(title = '3D Biplot',
scene = list(
xaxis = list(title = 'PC1'),
yaxis = list(title = 'PC2'),
zaxis = list(title = 'PC3')
))See it
Add labels
- This is a bit objective but you can just look at the biggest positive coefficient.
prc <- bind_cols(select(wino,variety),as.data.frame(pr_wine$x)) %>%
select(1:5) %>%
rename("points" = PC1) %>%
rename("fruit" = PC2) %>%
rename("tannin" = PC3) %>%
rename("finish" = PC4)
head(prc)# A tibble: 6 × 5
variety points fruit tannin finish
<chr> <dbl> <dbl> <dbl> <dbl>
1 Pinot_Gris -2.56 -0.453 -0.239 -1.41
2 Pinot_Noir -1.69 0.547 -2.42 -0.573
3 Pinot_Noir -0.274 0.728 0.0335 1.76
4 Pinot_Noir -1.52 -0.729 1.52 -1.36
5 Pinot_Noir -1.68 -1.50 0.195 -0.717
6 Pinot_Noir 1.39 0.213 2.17 0.411Density by variety (1&2)
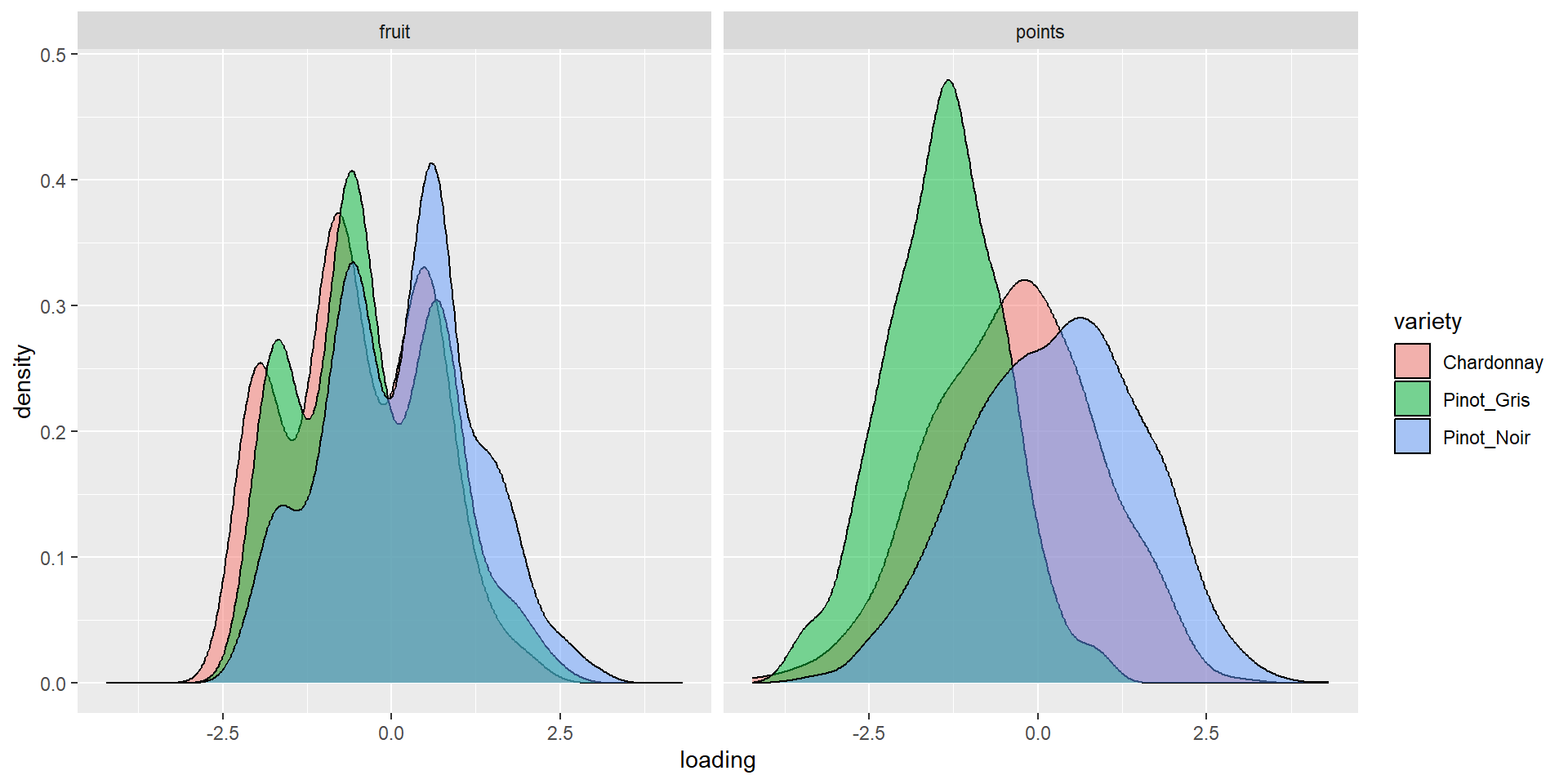
Density by variety (3&4)
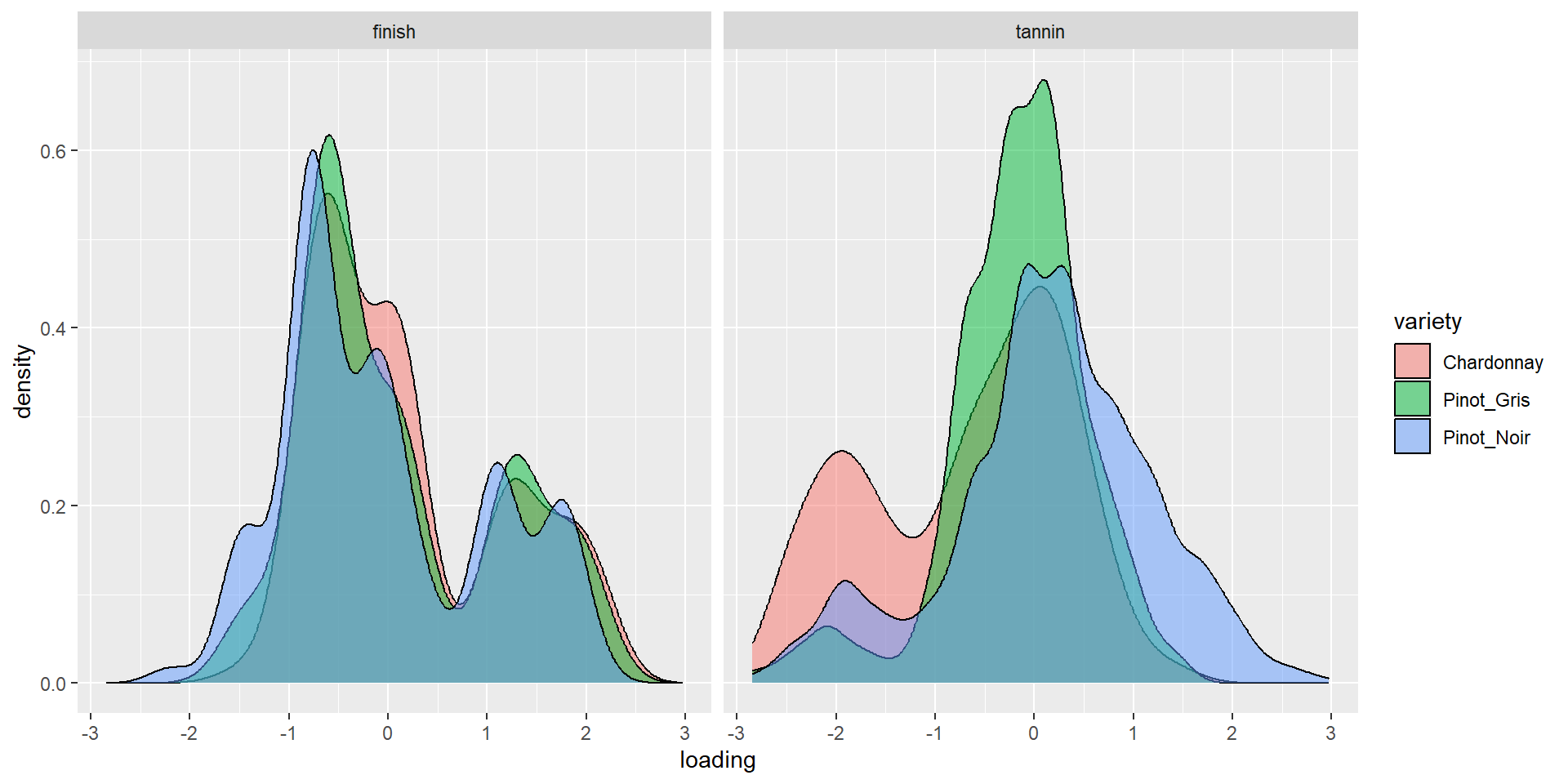
Aside
- This should be factored.
Use it
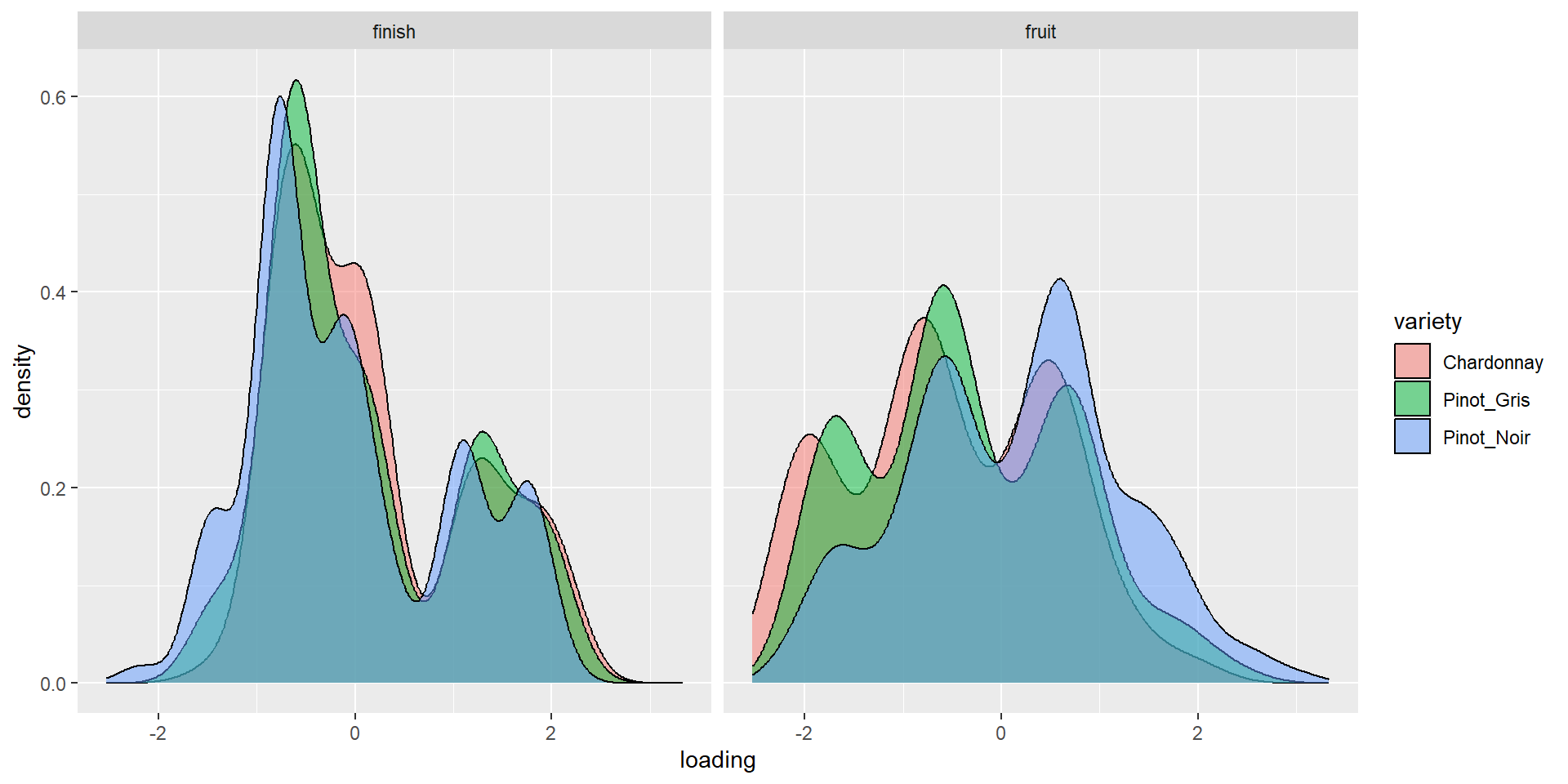
More Words
[1] "id" "variety" "price" "points" "description"- Why does changing “500” to “100” increase the number of words?
Run PCA

Highest loadings per factor
rownames_to_column(as.data.frame(pr_wine$rotation)) %>%
select(1:5) %>%
filter(abs(PC1) >= 0.25 | abs(PC2) >= 0.25 | abs(PC3) >= 0.25 | abs(PC4) >= 0.25) rowname PC1 PC2 PC3 PC4
1 months 0.09812814 0.30183350 0.282169826 -0.024209609
2 oak 0.06761861 0.36223058 0.274819423 -0.017899451
3 valley 0.04599743 0.02759857 0.001606928 0.444950984
4 pear -0.25009663 0.13746893 -0.132608010 0.003867835
5 french 0.09425237 0.33440763 0.276955369 -0.034348095
6 willamette 0.04915432 0.02738190 -0.001574066 0.440647855
7 points 0.26481521 0.19037913 -0.326062170 0.030618920
8 log_price 0.36816348 0.02944301 -0.064724155 -0.004236041- Price
- French Oak
- Also French Oak but not pears or points?
- Willamette Valley
Name it
Graph it
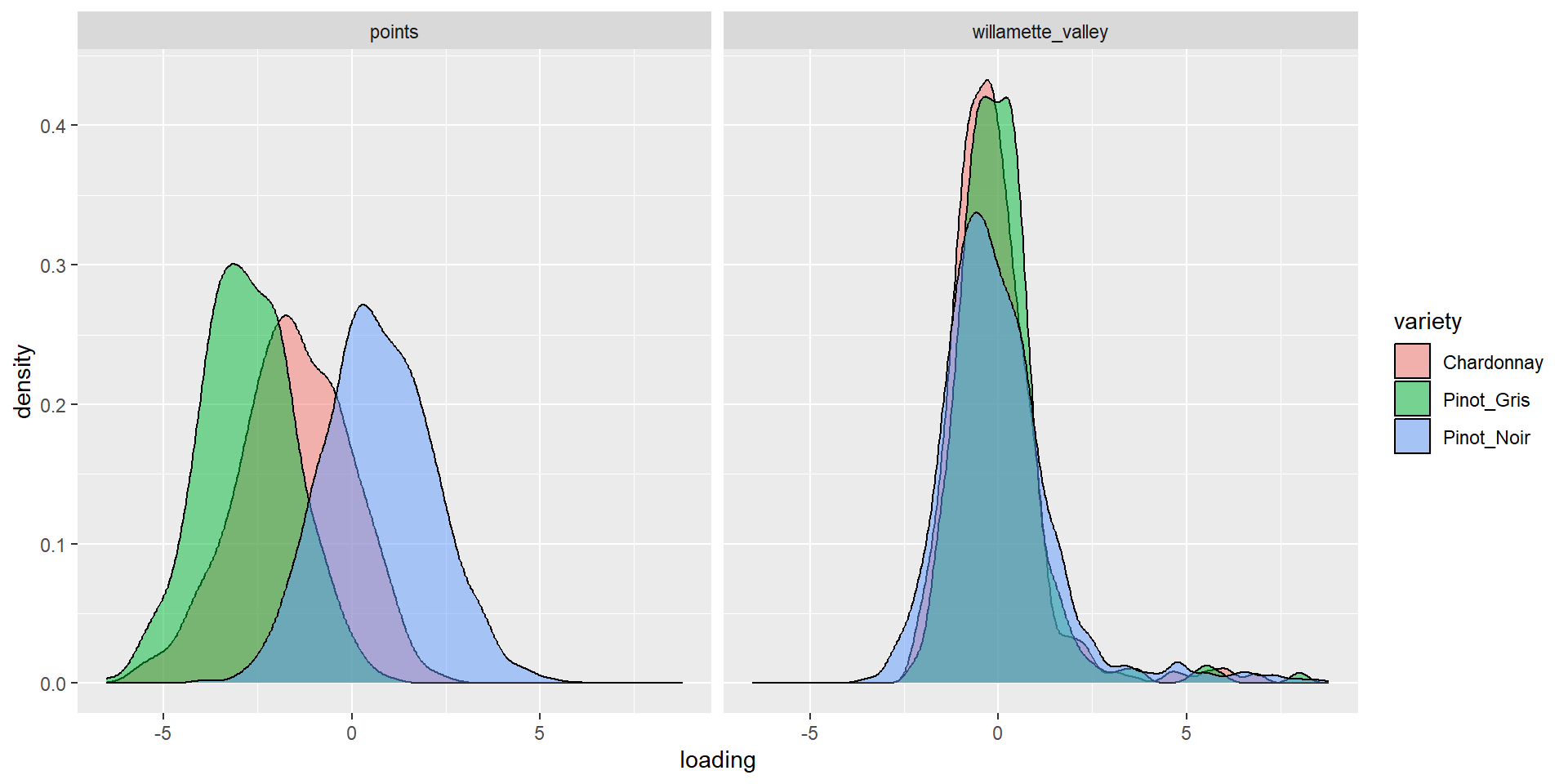
Graph more
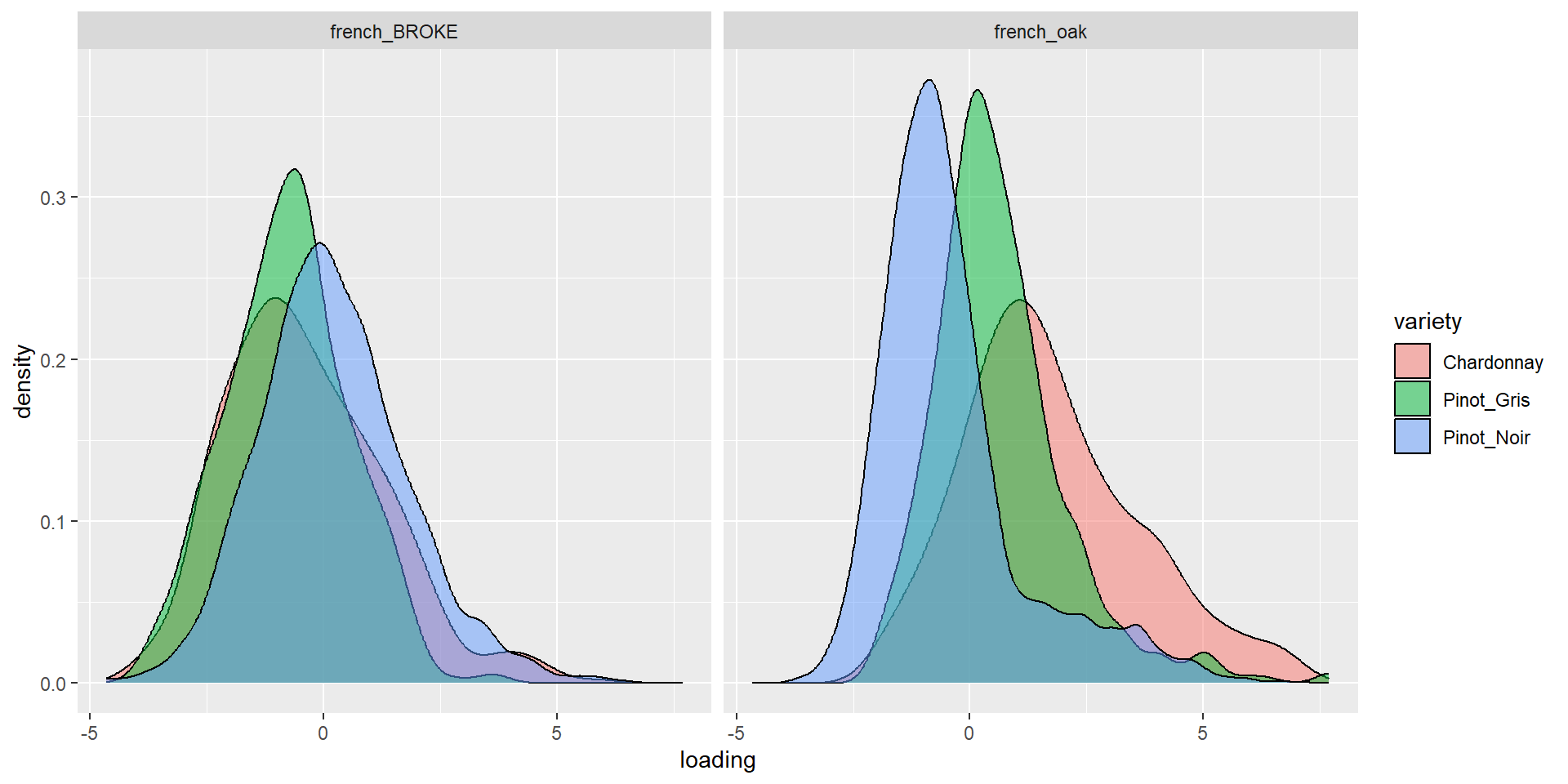
ML it
fit <- train(variety ~ .,
data = prc,
method = "naive_bayes",
metric = "Kappa",
trControl = trainControl(method = "cv"))
confusionMatrix(predict(fit, prc),factor(prc$variety))$overall['Kappa'] Kappa
0.6121089 - Not bad!
Planned Dinner
- Are we close??????
(Long) Exercise
- Load the bank data
- Run a principal component analysis on all the data except Churn
- Choose a number of factors based on a scree plot
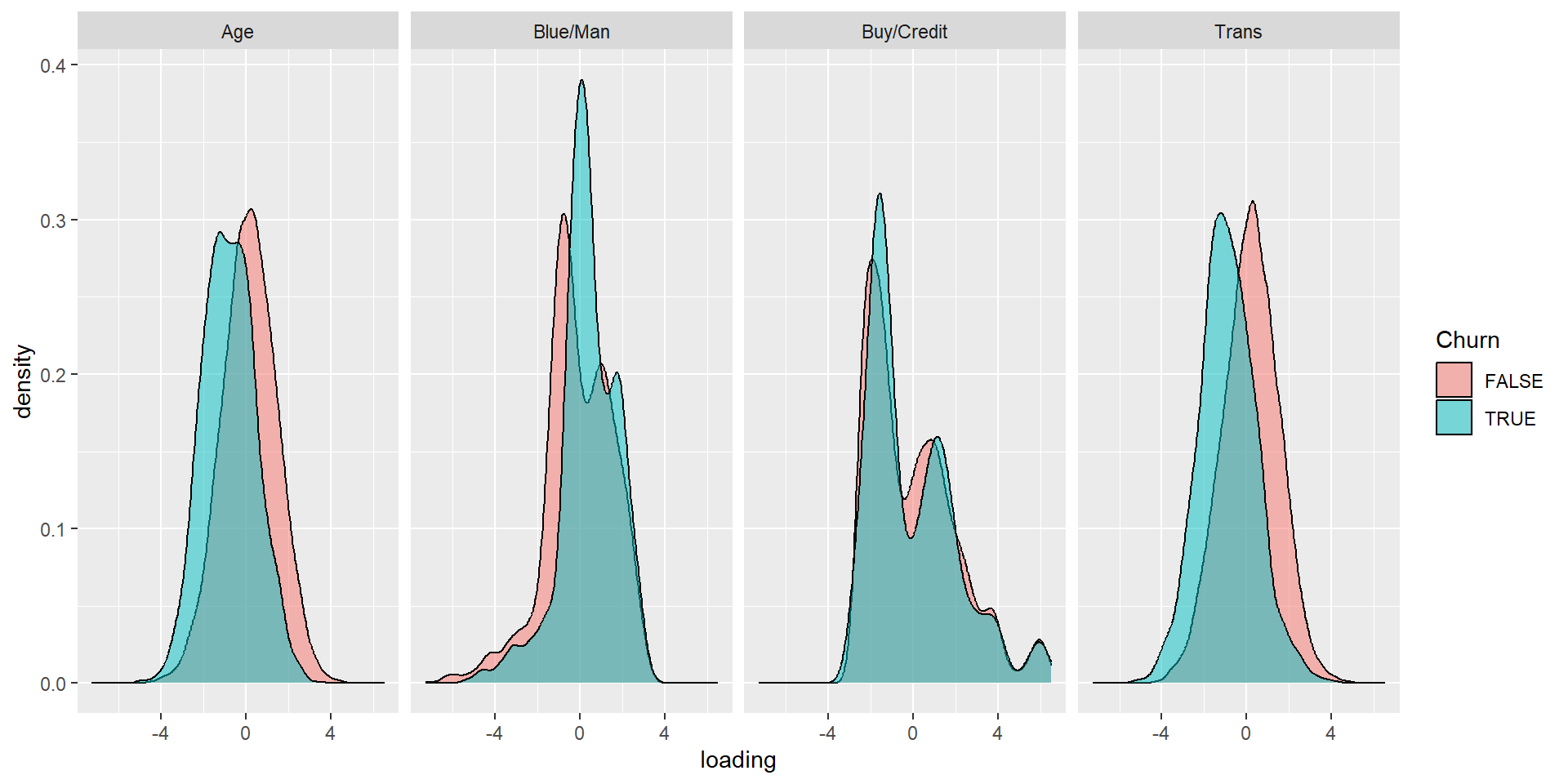
- name those factors, see whether you can interpret them
- Plot them against Churn using a density plot
Hendrik’s solution…
Hendrik’s solution…
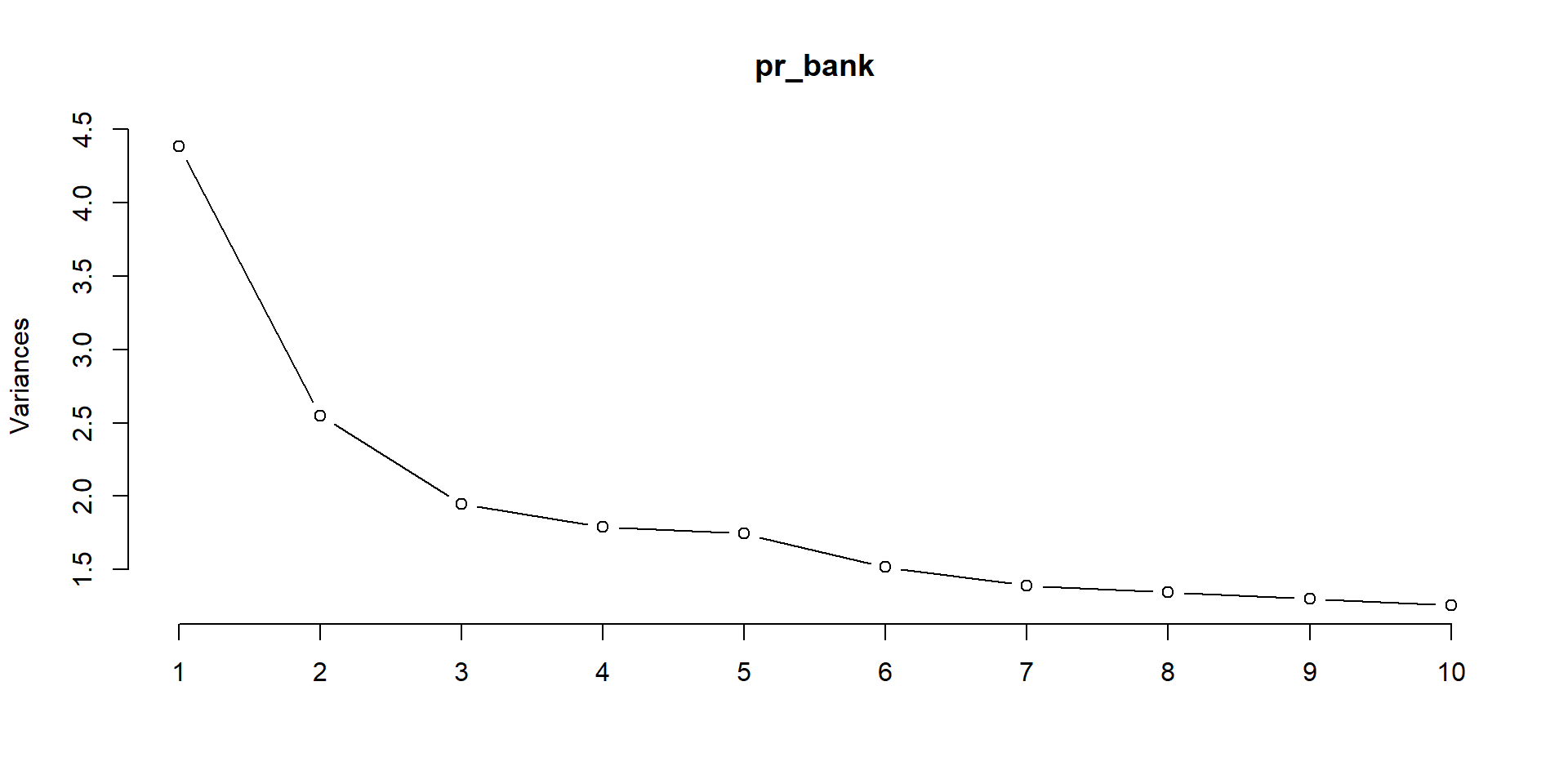
Hendrik’s solution…
rownames_to_column(as.data.frame(pr_bank$rotation)) %>% select(1:5) %>%
filter(abs(PC1) >= 0.35 | abs(PC2) >= 0.35 | abs(PC3) >= 0.35 | abs(PC4) >= 0.35) rowname PC1 PC2 PC3 PC4
1 Customer_Age -0.010813997 0.07575479 -0.4330194 0.443499234
2 Months_on_book -0.005699747 0.07134628 -0.4278530 0.439072869
3 Credit_Limit 0.413014059 -0.11846478 -0.1156352 -0.003755843
4 Avg_Open_To_Buy 0.416062573 -0.11834914 -0.1350212 -0.033748268
5 Total_Trans_Amt 0.100869534 -0.36992793 0.2898838 0.262444253
6 Total_Trans_Ct 0.044317988 -0.38057169 0.3032594 0.225273171
7 Gender_F -0.355263483 -0.32165703 -0.1903368 -0.067697739
8 Gender_M 0.355263483 0.32165703 0.1903368 0.067697739
9 Card_Category_Blue -0.257857151 0.35138834 0.1591349 -0.022247279Hendrik’s solution…
Hendrik’s solution…
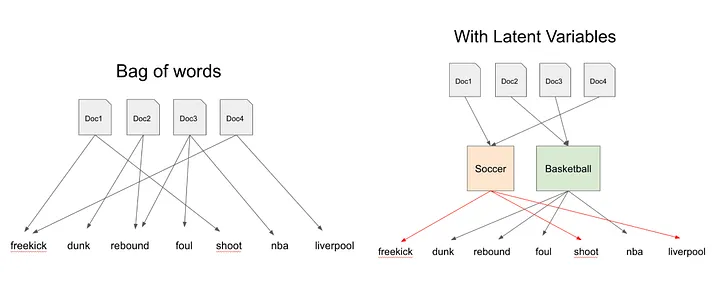
Topic Modeling
Latent Dirichlet allocation
- A common algorithms for topic modeling.
- Two principles:
- Every document is a mixture of topics.
- Every topic is a mixture of words.
Documents
We imagine that each document may contain words from several topics in particular proportions. For example, in a two-topic model we could say “Document 1 is 90% topic A and 10% topic B, while Document 2 is 30% topic A and 70% topic B.”
Topics
Every topic is a mixture of words. For example, we could imagine a two-topic model of American news, with one topic for “politics” and one for “entertainment.” The most common words in the politics topic might be “President”, “Congress”, and “government”, while the entertainment topic may be made up of words such as “movies”, “television”, and “actor”. Importantly, words can be shared between topics; a word like “budget” might appear in both equally.
Running a model
wine_dtm <- wine %>%
unnest_tokens(word, description) %>% anti_join(stop_words) %>%
filter(!(word %in% c("drink","vineyard","variety","price","points","wine","pinot","chardonnay","gris","noir","riesling","syrah"))) %>%
count(id,word) %>% cast_dtm(id, word, n) # DocumentTermMatrix
head(wine_dtm)<<DocumentTermMatrix (documents: 6, terms: 5836)>>
Non-/sparse entries: 100/34916
Sparsity : 100%
Maximal term length: 16
Weighting : term frequency (tf)Latent Dirichlet
Introduce two letters.
- Lowercase Beta, \(\beta\), per-topic-per-word probabilities
- Lowercase Gamma, \(\gamma\), per-document-per-topic probabilities
- More
- ctrl+f “beta” and “gamma”
- Use double quotes in the search bar!
Word-topic probabilities
Word-topic probabilities
top_terms <- topics %>%
group_by(topic) %>% top_n(10, beta) %>% ungroup() %>% arrange(topic, -beta)
top_terms# A tibble: 40 × 3
topic term beta
<int> <chr> <dbl>
1 1 flavors 0.0323
2 1 fruit 0.0230
3 1 tannins 0.00955
4 1 tart 0.00932
5 1 apple 0.00909
6 1 black 0.00882
7 1 red 0.00840
8 1 mix 0.00790
9 1 acidity 0.00761
10 1 chocolate 0.00736
# ℹ 30 more rowsWord-topic probabilities

Document-topic probabilities
What if we pivot wider?
wider <- function(df) {
df %>%
pivot_wider(id_cols = document,names_from = topic,values_from = gamma, names_prefix = "topic_") %>%
mutate(id=as.integer(document)) %>%
left_join(select(wine, id, variety)) %>%
select(-document, -id)
}
topics <- wider(topics)
head(topics)# A tibble: 6 × 5
topic_1 topic_2 topic_3 topic_4 variety
<dbl> <dbl> <dbl> <dbl> <chr>
1 0.254 0.247 0.247 0.251 Pinot_Gris
2 0.256 0.252 0.246 0.246 Pinot_Noir
3 0.253 0.249 0.249 0.249 Pinot_Noir
4 0.249 0.249 0.248 0.253 Pinot_Noir
5 0.252 0.252 0.251 0.245 Pinot_Noir
6 0.251 0.258 0.244 0.247 Pinot_NoirCan we model an outcome?
What if we used more topics?
# A tibble: 6 × 21
topic_1 topic_2 topic_3 topic_4 topic_5 topic_6 topic_7 topic_8 topic_9
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 0.0497 0.0501 0.0503 0.0497 0.0502 0.0496 0.0498 0.0499 0.0499
2 0.0502 0.0506 0.0503 0.0495 0.0503 0.0496 0.0498 0.0499 0.0496
3 0.0497 0.0499 0.0501 0.0500 0.0502 0.0497 0.0506 0.0496 0.0497
4 0.0501 0.0497 0.0499 0.0498 0.0504 0.0505 0.0503 0.0504 0.0498
5 0.0502 0.0498 0.0501 0.0503 0.0502 0.0501 0.0504 0.0500 0.0496
6 0.0499 0.0522 0.0502 0.0498 0.0501 0.0498 0.0502 0.0495 0.0495
# ℹ 12 more variables: topic_10 <dbl>, topic_11 <dbl>, topic_12 <dbl>,
# topic_13 <dbl>, topic_14 <dbl>, topic_15 <dbl>, topic_16 <dbl>,
# topic_17 <dbl>, topic_18 <dbl>, topic_19 <dbl>, topic_20 <dbl>,
# variety <chr>What if we used more topics?
Associated Press
Word-topic probabilities
# A tibble: 20 × 3
topic term beta
<int> <chr> <dbl>
1 1 aaron 3.41e- 5
2 2 aaron 1.02e- 5
3 1 abandon 3.94e- 5
4 2 abandon 2.87e- 5
5 1 abandoned 4.11e- 5
6 2 abandoned 1.50e- 4
7 1 abandoning 1.99e- 5
8 2 abandoning 6.77e- 6
9 1 abbott 3.44e- 5
10 2 abbott 7.32e-15
11 1 abboud 3.44e- 5
12 2 abboud 2.72e-45
13 1 abc 2.27e- 4
14 2 abc 6.49e- 5
15 1 abcs 6.80e- 5
16 2 abcs 1.57e- 5
17 1 abctvs 2.55e- 6
18 2 abctvs 3.15e- 5
19 1 abdomen 6.80e- 6
20 2 abdomen 3.65e- 5Top words by topic

Greatest beta differences
assoc_topics %>%
mutate(topic = paste0("topic", topic)) %>%
pivot_wider(names_from = topic, values_from = beta) %>%
head(20)# A tibble: 20 × 3
term topic1 topic2
<chr> <dbl> <dbl>
1 aaron 3.41e- 5 1.02e- 5
2 abandon 3.94e- 5 2.87e- 5
3 abandoned 4.11e- 5 1.50e- 4
4 abandoning 1.99e- 5 6.77e- 6
5 abbott 3.44e- 5 7.32e-15
6 abboud 3.44e- 5 2.72e-45
7 abc 2.27e- 4 6.49e- 5
8 abcs 6.80e- 5 1.57e- 5
9 abctvs 2.55e- 6 3.15e- 5
10 abdomen 6.80e- 6 3.65e- 5
11 abducted 1.78e-10 4.43e- 5
12 abduction 5.02e- 6 3.85e- 5
13 abductors 7.50e-29 2.95e- 5
14 abdul 5.18e- 7 2.89e- 5
15 abide 8.71e- 6 1.96e- 5
16 abilities 3.01e- 5 7.62e- 9
17 ability 1.65e- 4 8.21e- 5
18 ablaze 4.00e-13 6.39e- 5
19 able 3.18e- 4 3.93e- 4
20 abm 6.80e-13 3.44e- 5Greatest beta differences
Greatest beta differences
# A tibble: 30 × 4
term topic1 topic2 log_ratio
<chr> <dbl> <dbl> <dbl>
1 administration 0.00116 0.000796 -0.546
2 agency 0.000547 0.00118 1.12
3 agreement 0.00105 0.000707 -0.564
4 aid 0.000125 0.00122 3.29
5 air 0.000571 0.00160 1.49
6 american 0.00204 0.00158 -0.367
7 area 0.000142 0.00134 3.23
8 army 0.0000000371 0.00155 15.4
9 arrested 0.0000308 0.00100 5.02
10 asked 0.000926 0.00108 0.216
# ℹ 20 more rowsGreatest beta differences
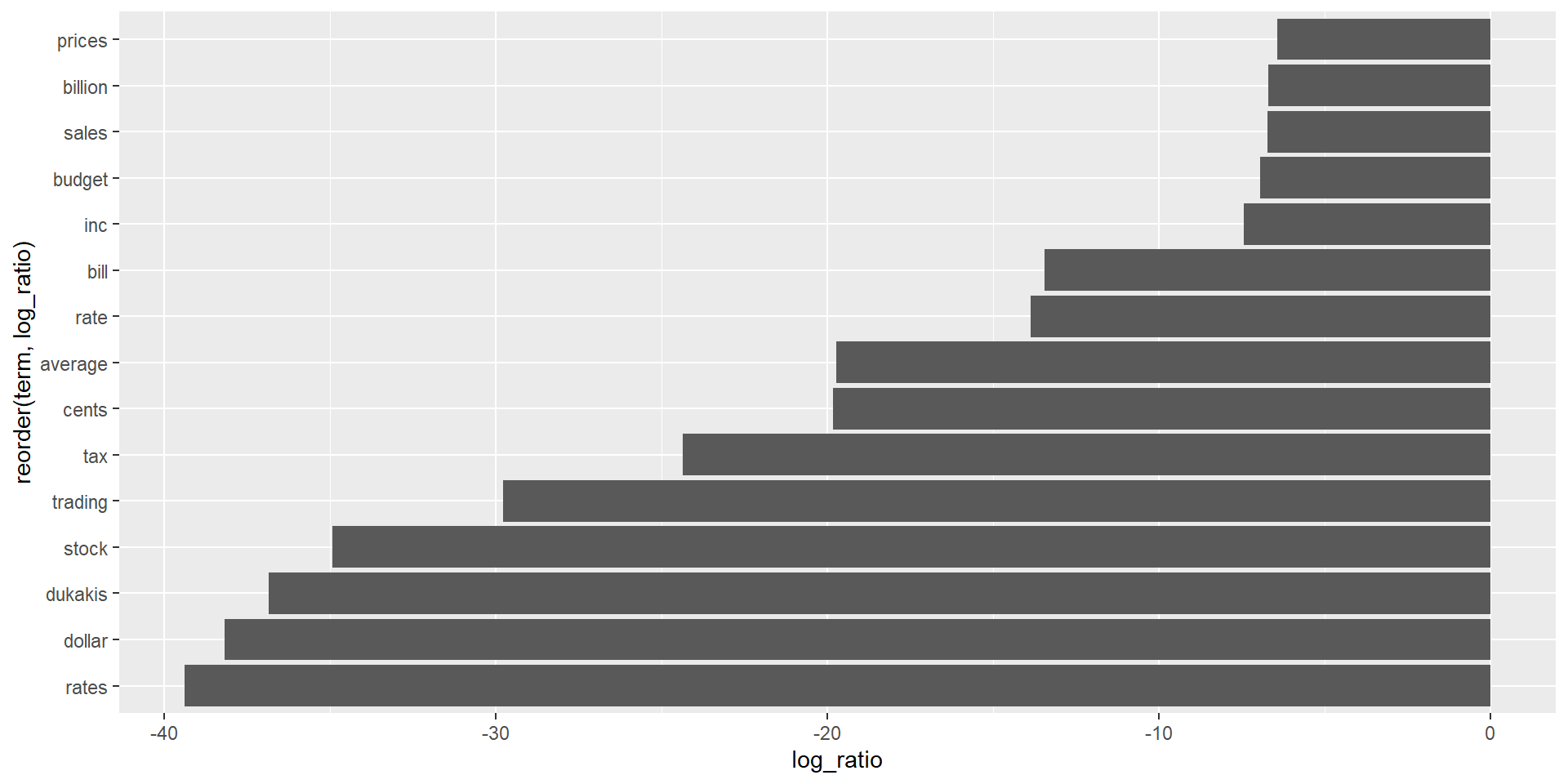
Greatest beta differences

Breakout Sessions
Group modeling project #2! Let’s get started.

